第七届广州核酸国际论坛
2019年11月7-8日 , 广州科学城
议题介绍
核酸科学一直都是全球生命科学与医学领域的核心研究方向,对推动全人类的精准医学、基因科学、细胞生物学、核酸药物和改善人类健康具有革命性的推动力和影响力,近年来核酸科学在诸多领域取得一系列原创性和突破性进展,成为最受学术与产业界瞩目的热点领域之一。
自2013年起举办的广州核酸国际论坛,历届论坛均由享誉世界的诺贝尔奖得主科学家领衔,由托马斯·切赫教授(1989年诺贝尔化学奖)和科学家克雷格·梅洛教授(2006年诺贝尔生理学或医学奖)担任大会执联主席,由三十多位国内外科学家担任科学委员会成员,旨在通过高端国际学术交流推动中国核酸科学和生物产业整体发展,促进全球范围内核酸科学学科发展,加快中国核酸产学研机构与国际顶尖学术及产业平台对接,促进中国同世界顶级科学家之间的对话。CNAF已经连续举办至第七届,演讲嘉宾均来自全球范围内顶级学术机构、科学团体或企业巨头,议题涵盖精准医学、生命科学、核酸药、生物标志物、非编码RNA等最新进展,论坛知名度与影响力得到国际学术界与产业界赞誉,成为中国核酸学术技术重要风向标。本届论坛主题如下:
1、全球核酸药最新发展
2、非编码RNA研究国际前沿
3、CRISPR基因编辑技术
4、RNA尖端技术与应用
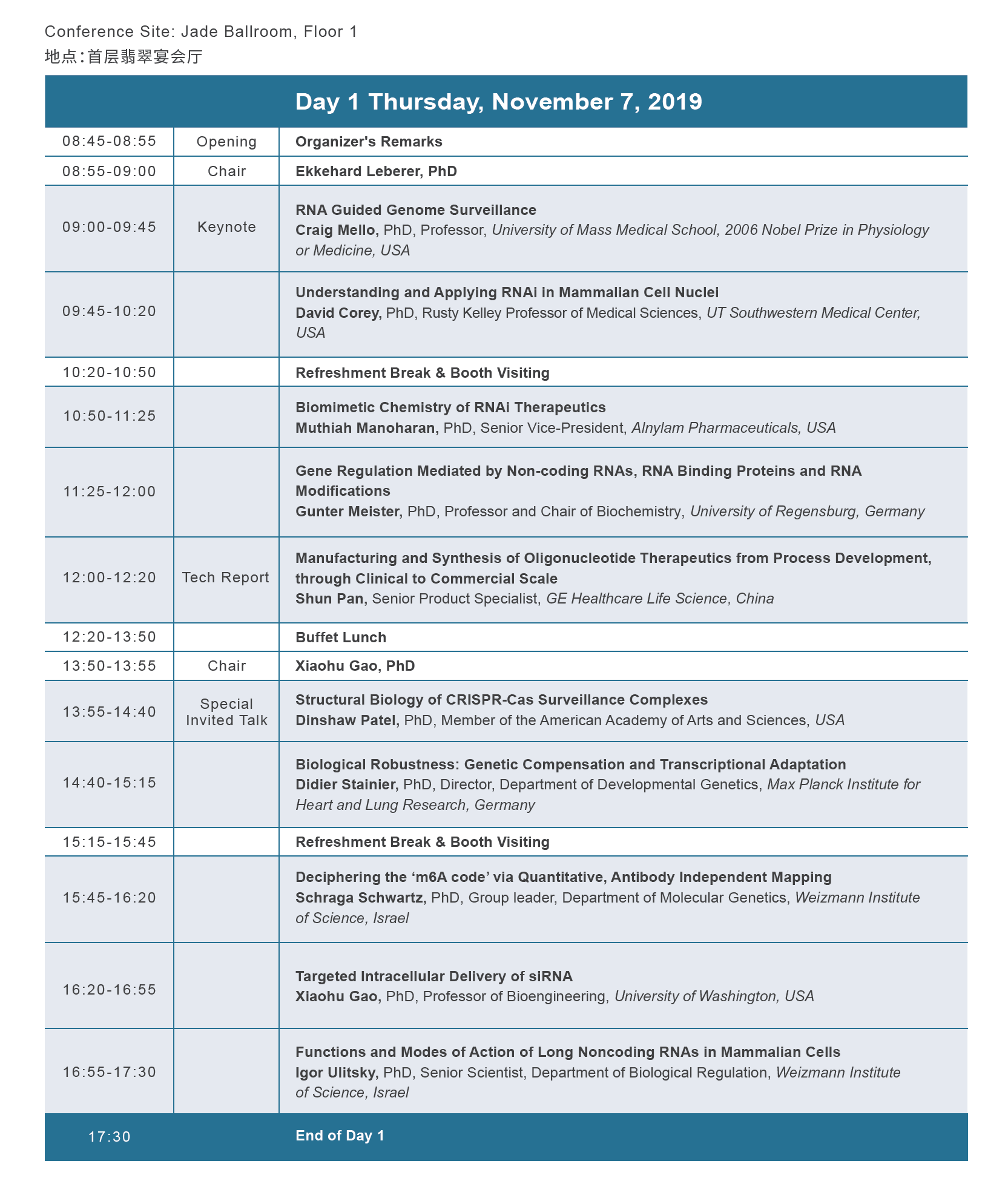
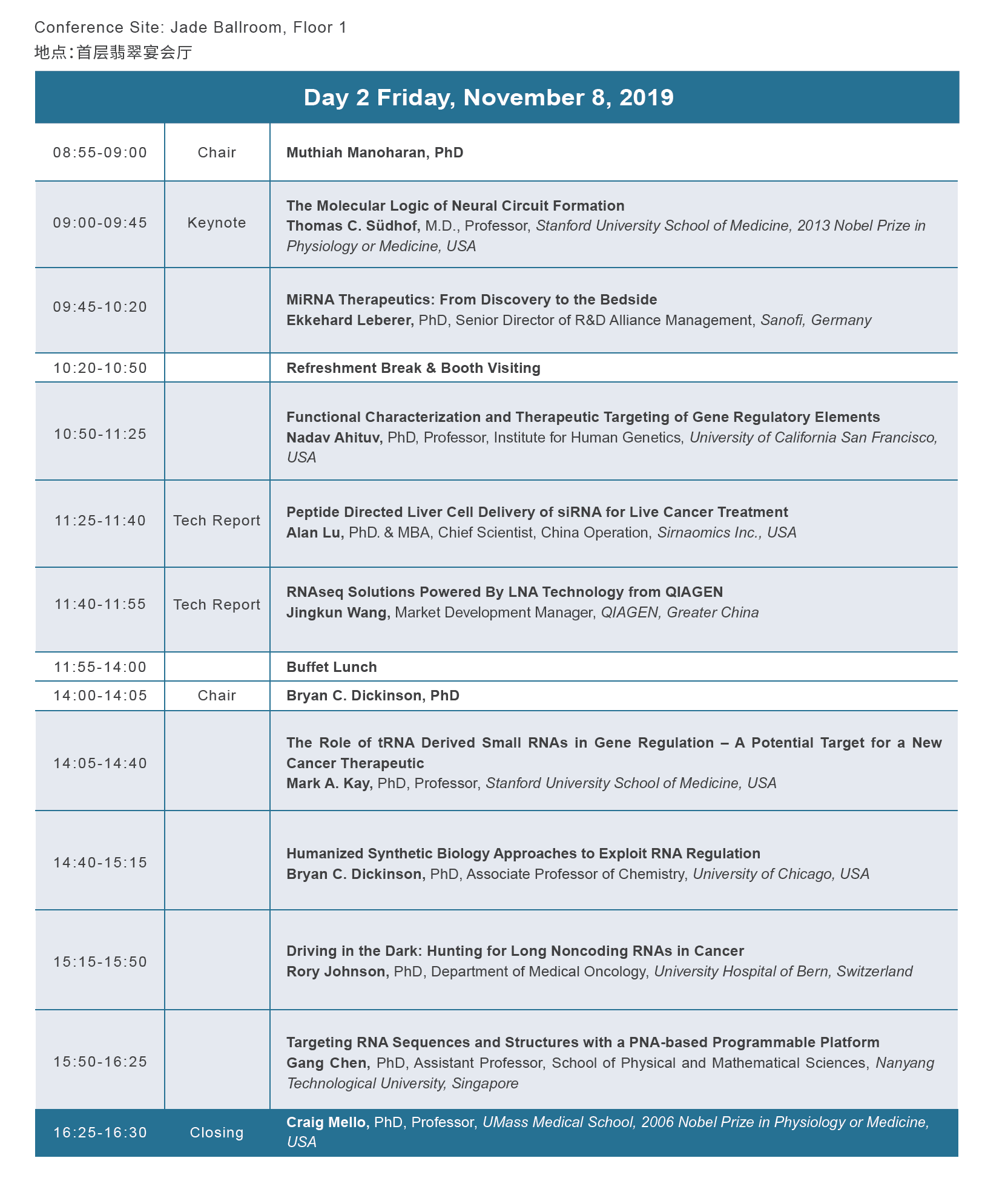
论坛议程
论坛赞助与合作媒体
金牌赞助

金牌赞助

银牌赞助

专项赞助










合作媒体





演讲嘉宾

Craig Mello, PhD
Professor, University of Massachusetts
Howard Hughes Medical Institute, USA
Nobel Prize in Physiology or Medicine (2006)
Dr. Mello’s lab uses the nematode C. elegans as a model system to study embryogenesis and gene silencing. His collaborative work with Dr. Andrew Fire led to the discovery of RNA interference (RNAi), for which they shared the 2006 Nobel Prize in Physiology or Medicine. Together they showed that when C. elegans is exposed to double-stranded ribonucleic acid (dsRNA), a molecule that mimics a signature of viral infection, the worm mounts a sequence-specific silencing reaction that interferes with the expression of cognate cellular RNAs. Using readily produced short synthetic dsRNAs, researchers can now silence any gene inorganisms as diverse as rice and humans. RNAi allows researchers to rapidly “knock out” the expression of specific genes and, thus, to define thebiological functions of those genes. RNAi also provides a potential therapeutic avenue to silence genes that cause or contribute to diseases.Dr. Mello received his BS degree in Biochemistry from Brown University in 1982, and PhD from Harvard University in 1990. From 1990 to 1994, he conducted postdoctoral research at the Fred Hutchinson Cancer Research Center in Seattle, WA. Now Dr. Mello is an Investigator of the Howard Hughes Medical Institute, the Blais University Chair in Molecular Medicine and Co-director of the RNA Therapeutics Institute at the University of Massachusetts Medical School.Besides the Nobel Prize, Dr. Mello’s work was recognized with numerous awards and honors, including the National Academy of Sciences Molecular Biology Award (2003), the Wiley Prize in Biomedical Sciences from Rockefeller University (2003), Brandeis University’s Lewis S. Rosnstiel Award for Distinguished Work in Medical Research (2005), the Gairdner Foundation International Award (2005), the Massry Prize (2005), the Paul Ehrlich and Ludwig Darmstaedter Award (2006), the Dr. Paul Janssen Award for Biomedical Research (2006), the Hope Funds Award of Excellence in Basic Research (2008). He is a member of the National Academy of Sciences, the American Academy of Arts and Sciences, and the American Philosophical Society.
Presentation Title
RNA Guided Genome Surveillance
Summary
Organisms experience a constant onslaught of invasive nucleic acids, such as viruses and transposable elements that cause genome instability and a broad range of diseases including cancer. The resulting arms race has driven the evolution of sophisticated nucleic acid-based immune pathways, such as RNA interference (RNAi) and prokaryotic CRISPR/CAS systems. The ability to recognize and respond to pathogenic nucleic acids, however, requires an equally sophisticated mechanism to avoid recognition and silencing of self-genes, or autoimmune silencing. Remarkably, this self/non-self dichotomy is regulated in C. elegans by opposing Argonaute pathways: the conserved Piwi pathway, which scans for and silences foreign genes, and the CSR-1 pathway, which recognizes and protects self-genes from Piwi-mediated autoimmune silencing. Both Argonautes and their small RNAs provide heritable epigenetic signals that confer stable modes of trans-generational gene regulation. We are exploring how CSR-1 interfaces with the conserved Piwi pathway, and the worm-specific AGO (WAGO) pathway to achieve whole-transcriptome surveillance of germline gene expression. We continue this journey into the surprising world of small RNAs with the hope that our research will shed new light on the mechanisms of transgenerational epigenetics inheritance and regulation of gene expression.

Thomas C. Südhof, M.D.
Avram Goldstein Professor and Professor of Molecular & Cellular Physiology, Neurosurgery, Psychiatry, and Neurology, Stanford University School of Medicine, USA
Investigator, Howard Hughes Medical Institute, USA
Dr. Südhof received his M.D. degree from the University of Göttingen, Germany, in 1982, after performing an internship in the Göttingen University Hospital. During his medical school training, he worked as an assistant scientist on his doctoral thesis at the Max-Planck-Institut für biophysikalische Chemie in Göttingen from 1978 to 1982, and was award a doctoral degree for this work also in 1982. After completing his medical and scientific training, Dr. Südhof became a postdoctoral fellow in 1983 in the laboratory of Drs. Michael S. Brown and Joseph L. Goldstein at the University of Texas Southwestern Medical School in Dallas, USA, where he cloned the LDL receptor gene and elucidated the cholesterol-dependent regulation of the expression of the LDL receptor. In 1986, Dr. Südhof founded his own laboratory and decided to completely switch fields to work in neuroscience. Over the next two decades, his laboratory first at Southwestern Medical School in Dallas and later at Stanford Medical School in Stanford described the molecular mechanisms underlying the calcium-dependent release of neurotransmitters at a synapse, work for which he was awarded the 2013 Albert Lasker Basic Medical Research Award and the 2013 Nobel Prize in Physiology or Medicine. In the last decade. Dr. Südhof’s work has shifted to a new basic problem in understanding the brain, which is the question of how neurons are wired into circuits via synaptic connections.
Presentation Title
The Molecular Logic of Neural Circuit Formation
Summary
Brain function is mediated by information processing in neural circuits. Circuits are formed by synapses that not only transfer information from one neuron to the next, but also process this information. Thus, synapses construct neural circuits, and represent the fundamental information processing units of the brain. Dr. Südhof’s current work focuses on the molecular mechanisms that establish a synaptic connection, the render it specific for a particular pair of neurons, and that endow a given synapse with characteristic properties. His presentation will outline key mechanisms that have been identified that explain these processes, and discuss their relevance to neuropsychiatric disorders.

Gunter Meister, PhD
Professor and chair of Biochemistry, University of Regensburg
Gunter Meister is professor and chair for Biochemistry at the University of Regensburg, Germany. He received his Master degree in Biology (Diploma) in 1999 from the University of Bayreuth, Germany. He obtained his PhD in 2002 from the Max-Planck-Institute of Biochemistry in Martinsried, and the Ludwig Maximilians University in Munich, Germany. From 2003 to 2005 he worked as postdoctoral fellow in the lab of Tom Tuschl at the Rockefeller University in New York, USA. In 2005, he started his own lab at the Max-Planck-Institute of Biochemistry as independent group leader. In 2009, he was appointed full professor and chair for Biochemistry at the University of Regensburg. His research focus is the biochemical analysis of small RNA-guided gene silencing pathways, long non-coding RNAs, RNA binding proteins and RNA modifications in mammals. He contributed to our understanding of the molecular basis of RNAi in human cells and also developed novel silencing approaches. He receives funding from the ERC (staring and consolidator grants) and received the research award of the Engelhorn foundation, the Schering young investigator award and the Bücher medal from FEBS.
Presentation Title
Gene regulation mediated by non-coding RNAs, RNA binding proteins and RNA modifications
Summary
Gene expression is not only regulated at the level of transcription, but also at many post-transcriptional steps. Various classes of non-coding RNAs have been identified that are vital for correct gene expression. This large RNA family includes microRNAs, lncRNAs or circular RNAs, which are generated by alternative splicing and can function as sponges for miRNAs or RNA-binding proteins. RNAs do not function as sole RNA molecules but are instead incorporated into RNA-protein complexes, in which at least one RNA-binding protein (RBP) contacts the RNA directly. Consequently, RBPs are essential for RNA metabolism and serve as regulatory hubs to coordinate the various RNA-guided processes. Mis-regulation of such processes is often associated with disease, a phenomenon that is generally underestimated. We have recently described how RBPs regulate miRNA expression on a co- or post-transcriptional level. In addition, we have started to investigate RNA modification pathways and their cross talks with other cellular RNA regulatory networks in human cells. I will present our recent progress on RBP functions including readers and writers of RNA base modifications. I will also present our efforts to generate robust and reliable tools for studying RNA modifications.

Igor Ulitsky, PhD
Senior Scientist
Department of Biological Regulation, Weizmann Institute of Science, Rehovot, Israel
Dr. Igor Ulitsky received his PhD in computational biology from Tel Aviv University in 2009, working with Prof. Ron Shamir on algorithms for combination of heterogenous biological data and discovery of functional models. He was then a postdoctoral fellow at Whitehead Institute for Biomedical Research in Cambridge, Massachusetts, working in the lab of Prof. David Bartel on the evolution and roles in embryonic development of long noncoding RNAs (lncRNAs). Since 2013, he is a group leader at the Weizmann Institute’s Department of Biological Regulation. His lab is combining experimental and computational biology to study the evolution, genomics, functions and modes of action of long noncoding RNAs in various mammalian systems, including stem cells, mouse models, and cancer cells. Dr. Ulitsky has been awarded a number of academic honors, including the Wolf Prize for Outstanding PhD Students (2008), the EMBO Long-Term Fellowship for postdoctoral research (2010–2011), and the Heineman Research Award (2018). He is an EMBO Young Investigator from 2016 and recipient of the ERC starting grant (2015). Lab website: https://www.weizmann.ac.il/Biological_Regulation/IgorUlitsky/
Presentation Title
Functions and modes of action of long noncoding RNAs in mammalian cells
Summary
It is now clear that many intergenic regions in eukaryotic genomes give rise to a range of processed and regulated transcripts that do not appear to code for functional proteins. A subset of these are long (>200 nt), capped, and polyadenylated RNAs transcribed by RNA polymerase II and collectively called long noncoding RNAs (lncRNAs). Given the growing number of lncRNAs implicated in human disease or required for proper development, fundamental questions that need to be addressed are: Which lncRNAs are functional? How is functional information encoded in the lncRNA sequence? Is this information interpreted in the context of the mature or the nascent RNA? We are tackling these questions by combination of experimental methods with a focus on lncRNA functions in early cell fate decisions and computational methods focused on lncRNA evolution.

Mark A. Kay, M.D., PhD
Dennis Farrey Family Professor, Head, Division of Human Gene Therapy, Departments of Pediatrics and Genetics at Stanford University School of Medicine
Professor Kay received his B.S. in physical sciences from Michigan State University, a Ph.D. in Developmental Genetics and M.D. from Case Western Reserve University. Afterwards, Dr. Kay pursued additional medical training and triple board certification in Pediatrics, Medical Genetics, and Inborn Errors of Metabolism at the Baylor College of Medicine. He was appointed to the faculty at the University of Washington in 1993 and moved to Stanford University in 1998.
Professor Kay’s lab made seminal contributions in the field of gene and nucleic acid based therapeutics. His early work on developing recombinant adeno-associated viral vectors led to the first clinical trial where such a vector was administered systemically into humans, and in which he held the original IND. His lab has made important contributions in the area of miRNA biogenesis, their mechanisms of action, and RNAi therapeutics. While his lab continues to work in these areas, his group was one of the first to promote the idea that tRNA derived small RNAs (tsRNAs) were not mere degradation products but rather molecules that play important roles in gene regulation. His lab has shown that one type of tsRNA functions in regulating ribosome biogenesis and is a potential target for treating hepatocellular carcinoma. Professor Kay has published over 250 papers, is currently the deputy editor of Human Gene Therapy, and serves on the editorial boards of other peer-reviewed publications.
Dr. Kay served on the Board of Directors of the Oligonucleotide Therapeutics Society for 10 years. He was on the founding board of the of the American Society of Gene and Cell Therapy, and served as its President in 2005-2006. He was the recipient of the society’s Outstanding Investigator Award in 2013. Dr. Kay received the E. Mead Johnson Award for Research in Pediatrics in 2000. He was elected to the American Society for Clinical Investigation in 1997 and the Association for American Physicians in 2010. He has organized many national and international conferences, including the first Gordon Conference dedicated to gene therapy. Professor Kay has and continues to serve on various commercial and academic boards. He is a scientific co-founder of Voyager Therapeutics, and a co-founder of LogicBio Therapeutics.
Presentation Title
The role of tRNA derived small RNAs in gene regulation – a potential target for a new cancer therapeutic
Summary
The function of tRNAs is not merely to provide the specific amino acids for a growing peptide chain during protein synthesis. tRNAs can be cleaved into various tRNA derived small RNAs. Prof. Kay will provide a brief introduction into this field, and describe the role and mechanism of how specific tsRNAs regulate ribosome biogenesis as well as why they represent potential targets for cancer treatment. He will provide an update on current approaches we are developing to help us identify biological targets of the myriad of various tsRNAs present in various cell types.

Muthiah Manoharan, PhD
Senior Vice-President, Alnylam Pharmaceuticals, USA
Dr. Muthiah (Mano) Manoharan serves as a Senior Vice President, Scientific Advisory Board Member, and a Distinguished Research Scientist at Alnylam Pharmaceuticals, Cambridge, Massachusetts, USA. In 2003, he was the first chemist hired at Alnylam. He and his team pioneered the discovery and development of the chemical modifications that make RNA interference-based human therapeutics possible. This work led to ONPATTRO (Patisiran), the first RNAi therapeutic approved by FDA in 2018. Dr. Manoharan has had a distinguished career as a world-leading chemist in the areas of oligonucleotide chemical modifications, conjugation chemistry, and delivery platforms (lipid nanoparticles, polymer conjugates, and complex-forming strategies). Dr. Manoharan and his research group demonstrated for the first time the human therapeutic applications of GalNAc-conjugated oligonucleotides at Alnylam, a platform that has revolutionized the nucleic acid-based therapeutics field with several compounds in the advanced clinical trials. He is an author of more than 215 publications (nearly 43,000 citations with an h-index of 94 and an i10-index of 367) and over 400 abstracts, as well as an inventor of over 225 issued U.S. patents. Prior to Alnylam, Dr. Manoharan worked at Ionis (formerly Isis) Pharmaceuticals and Lifecodes Corporation in the field of antisense oligonucleotide therapeutics. He received the M. L. Wolfrom Award from the American Chemical Society Carbohydrate Chemistry Division in 2007 for his contributions to this field. He has been recognized as the Lifetime Achievement Awardee of the Oligonucleotide Therapeutics Society for the year 2019.
Presentation Title
“Biomimetic Chemistry of RNAi Therapeutics”
Summary
• Recent Advances in RNAi Therapeutics
• Methods of Delivery
• Advances in GalNAc platform
• Addressing off-target effects
• Extra-hepatic Delivery
• Novel siRNA Chemistries

Ekkehard Leberer, PhD
Professor of Biochemistry
Senior Director of R&D Alliance Management, Sanofi, Germany
Scientific Managing Director of COMPACT Consortium, Innovative Medicines Initiative, Brussels, Belgium
Dr. Leberer received his Ph.D. in Biology at the University of Konstanz, Germany (1986), conducted post-doctoral training in molecular biology at the Banting and Best Institute of the University of Toronto, Canada. He then obtained the Habilitation for Professor of Biochemistry at the University of Konstanz, Germany (1992). From 1989-1998, he served as Senior Research Officer in genetics and genomics at the Biotechnology Research Institute, National Research Council of Canada, Montreal. He was also Adjunct Professor at McGill University, Montreal.
Since joining Hoechst Marion Roussel in 1998, Dr. Leberer carried out various managing roles in this company, Sanofi’s predecessor companies and Sanofi itself, including responsibilities in functional genomics, biological sciences and external innovation for oligonucleotide-based therapeutics. Since 2012, he is Global Alliance Manager for R&D at Sanofi, Frankfurt. In addition, from 2012-2018, he has been the Scientific Managing Director of the Innovative Medicines Initiative COMPACT Consortium on the delivery of biopharmaceuticals across biological barriers and cellular membranes, Brussels.
His research has focused on the molecular mechanisms of signal transduction and the role of signalling molecules in human diseases. He is the co-discoverer of the p21 activated protein kinase (PAK) family of cell signalling proteins and of novel virulence-inducing genes in pathogenic fungi. He is co-author of more than 60 publications in prestigious peer-reviewed journals including Nature and Science.
Presentation Title
MiRNA Therapeutics: From Discovery to the Bedside
Summary
The non-coding genome makes up 98.8% of the human genome. Most of this non-coding genome is transcribed into non-coding RNAs that may play an important role in cellular regulation in health and disease; these non-coding RNAs could be novel targets for future medicines.
MicroRNAs are short non-coding RNAs that regulate biochemical pathways and networks of pathways by the mechanism of RNA interference (RNAi). MicroRNA-21 has been implicated in multiple organs as a microRNA associated with fibrotic diseases and cancer.
The presentation will summarize the opportunities and challenges of developing microRNA-based drugs and will illustrate the successful generation of an anti-fibrotic microRNA-based therapeutic approach by targeting microRNA-21 with an antisense oligonucleotide (anti-miR-21). This microRNA-based drug is now in a phase 2 clinical trial for a fibrotic kidney disease called Alport Syndrome.

Bryan C. Dickinson, PhD
Associate Professor of Chemistry, University of Chicago, USA
Dr. Dickinson earned his B.S. in Biochemistry from the University of Maryland, College Park and his Ph.D. in Chemistry from the University of California at Berkeley for work performed with Professor Christopher Chang. His graduate work focused on the synthesis and application of small molecule fluorescent probes for the detection of hydrogen peroxide in living systems. Then, as a Jane Coffin Childs Memorial postdoctoral fellow with Professor David Liu at Harvard University, he developed new methods to rapidly evolve proteins to perform novel functions. Bryan joined the faculty at the University of Chicago in the Department of Chemistry in the Summer of 2014 and was promoted to Associate Professor in 2019. The Dickinson Group employs synthetic organic chemistry, molecular evolution, and protein design to develop molecular technologies to study chemistry in living systems. The group’s current primary research interests include: 1) how lipid modifications on proteins are controlled and regulate cell signaling, 2) developing new evolution technologies to reprogram and control biomolecular interactions, and 3) engineering protein-based systems to understand and exploit epitranscriptomic regulation. The motivating principle of the Dickinson Group is that our ability as chemists to create functional molecules through both rational and evolutionary approaches will lead to new breakthroughs in biology and biotechnology.
Presentation Title
Humanized synthetic biology approaches to exploit RNA regulation
Summary
Epitranscriptomic regulation controls information flow through the central dogma and provides unique opportunities for manipulating cell state at the RNA level. However, potential translational opportunities around RNA-targeting therapeutics are impeded by a lack of effective methods to target specific RNAs with effector proteins. The most common current approaches rely on large, microbially-derived systems that pose challenges when deployed as potential therapeutic strategies. To address this challenge, I will present the CRISPR/Cas-inspired RNA targeting system (CIRTS), a new protein engineering strategy for constructing programmable RNA regulatory systems constructed entirely from human protein parts. CIRTS represents a simple and generalizable approach to deliver a range of effector proteins to target transcripts, including nucleases, degradation machinery, translational activators, and base editors, which is also smaller than current Cas-based systems. The small size and human-derived nature of CIRTS provides a less-perturbative method for fundamental studies as well as a potential strategy to avoid immune issues when applied to epitranscriptomic therapies.

David Corey, PhD
Rusty Kelley Professor of Medical Sciences
Department of Pharmacology and Biochemistry
UT Southwestern Medical Center
Dr. Corey received his B.A. degree from Harvard in 1985 and then conducted his graduate work at University of California Berkeley Chemistry Department under the direction of Dr. Peter Schultz, receiving his Ph.D. in 1990. After postdoctoral work under the direction of Dr. Charles Craik at the University of California San Francisco Dr. Corey began his independent career at the University of Texas Southwestern Medical Center in 1992. Dr. Corey studies how chemically modified nucleic acids can be used to control gene expression. Ongoing project include activation of frataxin protein expression as a potential treatment for Friedreich’s Ataxia, understanding the role of RNAi in mammalian cell nuclei, and understanding the molecular causation and potential treatment of Fuchs’ Dystrophy, a disease caused by mutant trinucleotide repeat RNA.
Presentation Title
Understanding and applying RNAi in mammalian cell nuclei
Summary
RNAi is usually associated with the targeting of mRNA leading to inhibition of gene translation in the cytoplasm. We have observed that miRNAs and RNAi proteins like argonaute are also found in the nuclei of human cells. Duplex RNAs can be used to control gene transcription and splicing. We have appliced these insights from basic science to the development of synthetic nucleic acids that modulate expression of genes associated with disease. Friedreich’s Ataxia (FRDA) is an incurable genetic disorder caused by an mutant expansion of the trinucleotide GAA within an intronic FXN RNA that leads to reduced expression of frataxin (FXN) protein. Synthetic agents that increase levels of FXN protein might alleviate the disease. We demonstrate that introducing anti¬GAA duplex RNAs or single¬stranded locked nucleic acids (LNAs) into patient¬derived cells increases FXN expression to levels similar to analogous wild¬type cells. Our data identify synthetic nucleic acids as a strategy for restoring curative FXN levels.

Rory Johnson, PhD
Junior Group Leader at the Swiss National Centre for Competence in Research (NCCR) ‘RNA & Disease’.
Department of Medical Oncology, University Hospital of Bern, Switzerland.
Rory Johnson’s research program focusses on long noncoding RNAs, their basic biology and their roles in disease, using a combination of bioinformatics and CRISPR-Cas9 genome engineering. Johnson received his MSc in Physics from Imperial College, London, in 2000. He won a Wellcome Trust PhD scholarship at the University of Leeds (UK), where he wrote a thesis on transcriptional and post-transcriptional regulation of gene expression in the mammalian nervous system in 2006. His growing interest in lncRNAs led him first to the Genome Institute of Singapore, then to Roderic Guigo’s lab at the Centre for Genomic Regulation, Barcelona. Here, supported by a Ramon y Cajal fellowship, he worked with the GENCODE annotation project on methods to catalogue lncRNAs in the human and mouse genomes. During this same period, Johnson became interested in the potential of new CRISPR-Cas9 technology to address the great unanswered questions of the lncRNA field: which ones are functional, and which may be targeted in disease? This has been the objective of the Johnson laboratory at the University Hospital of Bern, composed of around 12 researchers, equally split between bioinformaticians and experimentalists. The lab has created a range of CRISPR tools for genome-wide functional screening of lncRNAs in cancer. They are also active members of GENCODE, the International Cancer Genome Consortium and the NCCR ‘RNA & Disease’. Finally, Johnson is involved in Knowledge and Technology Transfer, promoting collaborations between the pharmaceutical industry and NCCR research groups, to accelerate commercialization of RNA biology.
Presentation Title
Driving in the Dark: Hunting for Long Noncoding RNAs in Cancer
Summary
Conventional therapies for cancer, which target proteins using small molecules, are generally unable to provide effective and long lasting treatment. The recent discovery of tens of thousands of uncharacterized long noncoding RNAs (lncRNAs) in our genome, represents an exciting opportunity to develop new therapies, with enduring efficacy and low side-effects. We are working on a range of methods, both bioinformatic and experimental, to screen the vast space of lncRNAs for those with cancer-promoting activity. I will present an update on our latest results.

Dinshaw J. Patel, PhD
Member and Abby Rockefeller Mauze Chair in Experimental Therapeutics
Structural Biology Program Memorial Sloan-Kettering Cancer Center
USA
Dr. Dinshaw Patel received his PhD in Chemistry from New York University in 1968 for research in photochemistry. Following postdoctoral training in Biochemistry and Biophysics, he became a joined AT&T Bell Labs and spent the next 15 years undertaking NMR-based studies of the structure and dynamics of cyclic peptides, proteins and nucleic acids. He moved to Columbia University Medical School in 2004 where his group spent the next 8 years doing NMR-based research on DNA mismatches, bulges and junctions, on DNA triplexes and G-quadruplexes, and drug-DNA complexes. He was recruited in 1992 to the Memorial Sloan-Kettering Cancer Center to set up a Structural Biology program. His group’s research initially focused on NMR-based studies of covalent chiral carcinogen-DNA adducts, and complexes of antibiotics and peptides with natural and in vitro selected RNA targets. His laboratory has applied x-ray crystallography to projects on RNA-mediated gene regulation, with subsequent extension to histone-mark and DNA-mark mediated epigenetic regulation, to lipid transfer proteins, to nucleic acid pattern recognition receptors, and more recently cryoEM approaches to study CRISPR-Cas surveillance complexes and their inhibition by anti-CRISPR proteins. His group has complemented their structural efforts with functional studies to deduce mechanistic insights into the biological systems of interest. Dr. Patel is a Member of the National Academy of Sciences and the American Academy of Arts and Sciences.
Presentation Title
Structural Biology of CRISPR-Cas Surveillance Complexes
Summary
The role of RNA in information transfer and catalysis highlights its dual functionalities. Our ongoing research on RNA-mediated gene regulation has focused on CRISPR-Cas pathways involving ternary complexes of recently identified single-subunit Cas12 surveillance complexes with bound guide RNA and target DNA.
The lecture component on multi-subunit CRISPR-Cas complexes will focus on the type I-A Csy and III-A Csm systems. In the type III-A Csm system, we shall address mechanisms of target RNA binding and cleavage, as well as target RNA-activated ssDNA cleavage in the HEPN pocket and cyclic oligoadenylate (cOA) formation from ATP in the Palm pocket. We will also highlight a timer mechanism whereby successive nicks of bound cOA within the CARF domain regulate the trans-acting Csm6 RNase activity of the adjacent HEPN domain.
We have also investigated the impact of anti-CRISPR proteins in suppressing single- and multi-subunit CRISPR-Cas host defense pathways.
The above studies were undertaken by postdoc Hui Yang (x-ray studies on single-subunit Cas 12) and in collaboration with the Sriram Subramaniam group at NCI (cryo-EM studies on multi-subunit type 1 Csy) and postdoc Ning Jia (x-ray and cryo-EM studies on multi-subunit type III-A Csm).

Xiaohu Gao, PhD
Professor of Bioengineering, University of Washington, USA
Dr. Gao received his B.S. degree from Nankai University in China (1998), his Ph.D. degree in chemistry from Indiana University, Bloomington (2004), and his postdoctoral training from the Department of Biomedical Engineering at Georgia Tech and the Winship Cancer Institute at Emory University. He has been a faculty member in the Department of Bioengineering at the University of Washington since 2005. His research interests include cancer nanotechnology, molecular engineering, molecular imaging, and drug delivery. He has published >90 peer-reviewed journal articles.
Presentation Title
Targeted intracellular delivery of siRNA
Summary
Biomacromolecular agents such as DNA, RNA, proteins, and peptides are often superior in target binding specificity and easier to design compared to small-molecule drugs. A fundamental limitation for these large, highly water-soluble molecules, however, is their inability to access the intracellular space where the vast majority of biological activities take place. In this talk, I will discuss recent progresses we made on targeted intracellular delivery of biologics. This work has the potential to expand the target space for both diagnosis and therapy.

Didier Stainier, PhD
Director, Department of Developmental Genetics
Max Planck Institute for Heart and Lung Research, Germany
Dr. Didier Stainier is the director of the Department of Developmental Genetics at the Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany. He received his Ph.D. in Biochemistry and Biophysics from Harvard University where he studied the cellular basis of axon guidance and target recognition in the developing mouse brain with Walter Gilbert. After a postdoc with Mark Fishman at Massachusetts General Hospital (Boston) where he initiated the studies on zebrafish cardiac development, he spent close to 20 years at the University of California San Francisco, expanding his research to investigate questions of cell differentiation, tissue morphogenesis, organ homeostasis and function, as well as organ regeneration, in the zebrafish cardiovascular system and endodermal organs. In 2012, he moved to the Max Planck Institute where he continues to utilize both forward and reverse genetic approaches to investigate cellular and molecular mechanisms of developmental processes during vertebrate organ formation, in both zebrafish and mouse. His lab also studies mechanisms of genetic compensation in zebrafish, mouse, C. elegans and yeast.
Presentation Title
Biological Robustness: genetic compensation and transcriptional adaptation
Summary
Genetic robustness, or the ability of an organism to maintain fitness in the presence of harmful mutations, can be achieved via protein feedback loops. Previous work has suggested that organisms may also respond to mutations by transcriptional adaptation, a process by which related gene(s) are upregulated independently of protein feedback loops. However, the prevalence of transcriptional adaptation and its underlying molecular mechanisms are unknown. Here, by analyzing several models of transcriptional adaptation in C. elegans, zebrafish and mouse, we uncover a requirement for mutant mRNA degradation. Alleles that fail to transcribe the mutated gene do not exhibit transcriptional adaptation, and these alleles give rise to more severe phenotypes than alleles displaying mutant mRNA decay. Transcriptome analysis in alleles displaying mutant mRNA decay reveals the upregulation of a substantial proportion of the genes that exhibit sequence similarity with the mutated gene’s mRNA, suggesting a sequence-dependent mechanism. Furthermore, using the C. elegans model, we uncover a requirement for factors known to be involved in small RNA maturation and transport into the nucleus including Argonaute proteins and DICER, and confirm these findings in mouse models. These results have implications for our understanding of disease-causing mutations, and will help in the design of mutant alleles with minimal transcriptional adaptation-derived compensation.

Schraga Schwartz, PhD
Group leader, Department of Molecular Genetics, Weizmann Institute of Science
Dr. Schwartz received his B.Sc. from Tel Aviv University in Israel in 2006, where he then also conducted his PhD work in the field of RNA splicing under the supervision of Prof. Gil Ast, obtaining his PhD in 2010. Dr. Schwartz then conducted his post-doctorate at the Broad Institute of MIT & Harvard under the joint supervision of Aviv Regev and Eric Lander, where his work focused on mRNA modifications, and he established his independent laboratory at the Weizmann Institute in 2015. Dr. Schwartz has pioneered some of the key approaches for systematically detecting and quantifying RNA modifications at a transcriptome-wide scale. His lab bridges experimental and computational approaches, which jointly allowed the development of the first approach for systematically mapping N6-methyladenosine (m6A), pseudouridine, and the first approach for obtaining single-nucleotide resolution mappings of N1-methyladenosine (m1A). His lab aims to unravel the functions and mechanisms of actions through which diverse RNA modification modulate the fate of RNA, and understand how this in turns shapes cellular decision-making.
Presentation Title
Deciphering the ‘m6A code’ via quantitative, antibody independent mapping
Summary
N6-methyladenosine (m6A) is the most abundant modification on mRNA and is implicated in critical roles in development, physiology, and disease. A major limitation has been the inability to quantify m6A stoichiometry and the lack of antibody-independent methodologies for interrogating m6A. We developed MAZTER-seq for systematic quantitative profiling of m6A at single-nucleotide resolution at 16%-25% of expressed sites, building on differential cleavage by an RNase. MAZTER-seq permits validation and de novo discovery of m6A sites, calibration of the performance of antibody-based approaches, and quantitative tracking of m6A dynamics in yeast gametogenesis and mammalian differentiation. We discover that m6A stoichiometry is “hard coded” in cis via a simple and predictable code, accounting for 33%-46% of the variability in methylation levels and allowing accurate prediction of m6A loss and acquisition events across evolution. MAZTER-seq allows quantitative investigation of m6A regulation in subcellular fractions, diverse cell types, and disease states.

Nadav Ahituv, PhD
Professor
Department of Bioengineering and Therapeutic Sciences
Institute for Human Genetics
University of California San Francisco, USA
Dr. Nadav Ahituv is a Professor in the Department of Bioengineering and Therapeutic Sciences and the Institute for Human Genetics at the University of California, San Francisco. He received his PhD in human genetics from Tel-Aviv University working on hereditary hearing loss. He then did his postdoc, specializing in functional genomics, in the Lawrence Berkeley National Laboratory and the DOE Joint Genome Institute. His current work is focused on identifying gene regulatory elements and linking nucleotide variation within them to various phenotypes including morphological differences between species, drug response and human disease. In addition, his lab is developing massively parallel reporter assays (MPRAs) that allow for high-throughput functional characterization of gene regulatory elements and the use of gene regulatory elements as therapeutic targets.
Presentation Title
Functional characterization and therapeutic targeting of gene regulatory elements
Summary
Nucleotide variation in gene regulatory elements is a major determinant of phenotypes including morphological diversity between species, human variation and human disease. Despite continual progress in the cataloging of these elements, little is known about the code and grammatical rules that govern their function. Deciphering the code and their grammatical rules will enable high-resolution mapping of regulatory elements, accurate interpretation of nucleotide variation within them and the design of sequences that can deliver molecules for therapeutic purposes. To this end, we are using massively parallel reporter assays (MPRAs) to simultaneously test the activity of thousands of gene regulatory elements in parallel. By designing MPRAs to learn regulatory grammar or to carry out saturation mutagenesis of every possible nucleotide change in disease causing gene regulatory elements, we are increasing our understanding of the phenotypic consequences of gene regulatory mutations. Regulatory elements can also serve as therapeutic targets. To highlight this role, we used CRISPR/Cas9 activation (CRISPRa) of regulatory elements to rescue a haploinsufficient disease (having ~50% dosage reduction due to having only one functional allele) in vivo. By targeting the Sim1 promoter or its 270kb distant hypothalamic enhancer, we were able to rescue the haploinsufficient obesity phenotype in Sim1 heterozygous mice, both using a transgenic and adeno-associated virus approach. Our results highlight how regulatory elements could be used as therapeutic targets to treat numerous altered gene dosage diseases.

Gang Chen, PhD
Assistant Professor, Division of Chemistry and Biological Chemistry, School of Physical and Mathematical Sciences, Nanyang Technological University, Singapore
Dr. Gang CHEN received his B.S. degree in Chemistry at the University of Science and Technology of China (USTC) in 2001. He did his Ph.D. studies with Prof. Douglas TURNER in the Department of Chemistry at the University of Rochester. His Ph.D. work involved thermodynamic and NMR studies of RNA internal loops. A better understanding of the sequence dependence of thermodynamics for RNA structures will improve the accuracy of the RNA secondary structure prediction programs such as MFOLD and RNAstructure. He earned his Ph.D. in 2005. He was a postdoctoral fellow in Prof. Ignacio TINOCO’s lab in the Department of Chemistry at the University of California, Berkeley from January 2006 to June 2009. His research in Tinoco lab was on single-molecule mechanical unfolding and folding of RNA pseudoknots by laser optical tweezers, which provided new insights into ribosomal reading-frame regulation by cis-acting mRNA structures. He was a Research Associate in Prof. David MILLAR’s lab in the Department of Molecular Biology at The Scripps Research Institute working on HIV-1 Rev-RRE assembly using single-molecule fluorescence techniques. In July 2010, he joined the faculty in the Division of Chemistry and Biological Chemistry at Nanyang Technological University in Singapore.
Dr CHEN’s research group in Singapore has been working on probing and targeting RNA structures. By nano-manipulation using optical tweezers in combination with other biophysical methods, the group has revealed at the single-molecule level the contributions of molecular recognition interactions such as a single hydrogen bond in a base pair or base triple to RNA folding and unfolding and characterized the effects of a single proton binding. The lab has unveiled the correlations (i) between mRNA mechanical properties and mRNA structure induced translational reading frame shifting and (ii) between mechanical properties of tau pre-mRNA splice site hairpin structures and pre-mRNA alternative splicing activities. In addition, the lab is developing a molecular recognition platform based on chemically modified Peptide Nucleic Acids (PNAs) for the targeting of RNA structures in a sequence- and structure-specific manner. Specifically, the lab programs the dsRNA-binding PNAs (dbPNAs) with a new four-letter chemical code (T, L or R, E or S, and Q) for the recognition of RNA Watson–Crick duplexes, through the formation of major-groove triples. The lab has demonstrated that short (e.g., 10-mer) PNAs containing L, R, E, S, or Q show enhanced sequence-specific recognition of RNA base pairs in dsRNAs with significantly weakened binding to ssRNAs or dsDNAs at near-physiological conditions. The cell culture studies show that PNAs conjugated with small molecules are bioactive in the targeting of viral and cellular RNAs. The lab is further developing the dbPNAs platform as useful diagnostic tools and therapeutic drugs to fight against viral infections, cancers, and neurodegenerative diseases.
Presentation Title
Targeting RNA sequences and structures with a PNA-based programmable platform
Summary
RNAs perform a diverse array of catalytic and regulatory functions in viruses and cells and are becoming increasingly important disease biomarkers and drug targets. RNA structures are mainly stabilized by base paired double-stranded (ds) stem regions. Together with single-stranded (ss) loop regions, RNAs can fold into complex secondary and tertiary structures, facilitating the molecular recognition of RNA structures by small molecules and peptides/proteins. Currently, programmable RNA structure-specific and tight-binding ligands are relatively unexplored. Peptide nucleic acid (PNA) is characterized by a neutral, peptide-like backbone, with superior chemical stability and strong binding to complementary RNA/DNA sequences. Importantly, PNAs conjugated with cell-penetrating moieties show promising bioactivities in animal models. In this presentation, I will present our results on the synthesis and biophysical characterization of the novel chemically-modified dsRNA-binding PNAs (dbPNAs), which show selective recognition of dsRNAs over ssRNAs and dsDNAs in a sequence-specific manner. dbPNAs and traditional antisense PNAs (asPNAs) can be combined to serve as useful dsRNA-ssRNA junction-specific molecular glues for the probing and targeting of many biologically and medically important RNA structures in transcriptomes. I will discuss the applications of the PNAs platform in targeting a miRNA hairpin precursor, a tau pre-mRNA splice site hairpin structure, and an influenza viral RNA structure.
论坛起源于广州核酸国际论坛,在政府及各界支持下已经连续举办了七届,历届大会报告人汇集诺贝尔奖得主在内的全球顶级科学家、学者和企业家,论坛聚焦精准医学、生命科学、基因治疗、核酸药开发等最新热点进展,分享来自全球范围内核酸研究及应用的重大成果和进展。广州核酸国际论坛(CNAF)的知名度与影响力也受到国际学术界与企业界的高度赞誉,被誉为中国核酸研究与产业化的重要风向标。

